Getting started with OpenMOLE
When you build a model, you need to understand and/or optimize some parameters and to understand their effect on the dynamics of your model. OpenMOLE has been built for that!
First, you embed your model in OpenMOLE (5 minutes),
Then, you pick one of the distributed calibration genetic algorithms provided by OpenMOLE (5 minutes) and
Finally, you launch the model exploration on a distributed execution environment with thousands of machines (1 minute once authentication is set up). To summarize, you can design large scale model exploration reusing legacy code and advanced numeric methods in approximately 10 minutes.
First, you embed your model in OpenMOLE (5 minutes),
Then, you pick one of the distributed calibration genetic algorithms provided by OpenMOLE (5 minutes) and
Finally, you launch the model exploration on a distributed execution environment with thousands of machines (1 minute once authentication is set up). To summarize, you can design large scale model exploration reusing legacy code and advanced numeric methods in approximately 10 minutes.
You can try OpenMOLE online using the OpenMOLE demonstration site.
OpenMOLE requires that Java version 8 or above is set as the default java on your computer. To make sure of that you can check our FAQ.
Then just extract the archive, available here, somewhere and you're done! OpenMOLE is installed and works out of the box!
Once installed, you can launch OpenMOLE by executing the openmole file in the installation directory (it is called openmole.bat for windozers). It will bring up you web browser. OpenMOLE supports Chrome and Firefox. So if you are using another default web browser please copy paste the OpenMOLE url (http://localhost:port) in one of these browser.
You should see something like this :
Then just extract the archive, available here, somewhere and you're done! OpenMOLE is installed and works out of the box!
Once installed, you can launch OpenMOLE by executing the openmole file in the installation directory (it is called openmole.bat for windozers). It will bring up you web browser. OpenMOLE supports Chrome and Firefox. So if you are using another default web browser please copy paste the OpenMOLE url (http://localhost:port) in one of these browser.
 .
.
OpenMOLE proposes several launching options. Execute
openmole -h to list all the available options.
To run OpenMOLE on a remote machine and connect to using a web browser you should run:
openmole --remote --port portnumber. The first time you launch it, it asks you for a password. Then you can remotely connect to OpenMOLE using the url https://remotemachine:portnumber - https:// is important. When you connect it asks you for the password you've entered previously.
OpenMOLE proposes a headless mode for running scripts. You can enable it thanks to the -s option:
In that case OpenMOLE still asks for your preferences cyphering password. To provide it at launch time use the -pw option:
./openmole -s /path/to/you/mole/script
In that case OpenMOLE still asks for your preferences cyphering password. To provide it at launch time use the -pw option:
./openmole -s /path/to/your/mole/script --password password. A better practice is to write this password in a file readable by OpenMOLE only and use ./openmole -s /path/to/your/mole/script --password-file password.txt.
Let's explore (i.e. iterate within its domain) one variable, multiply it by 2 and then displays each result. The computation is executed in a multi-threaded environment on the local machine.
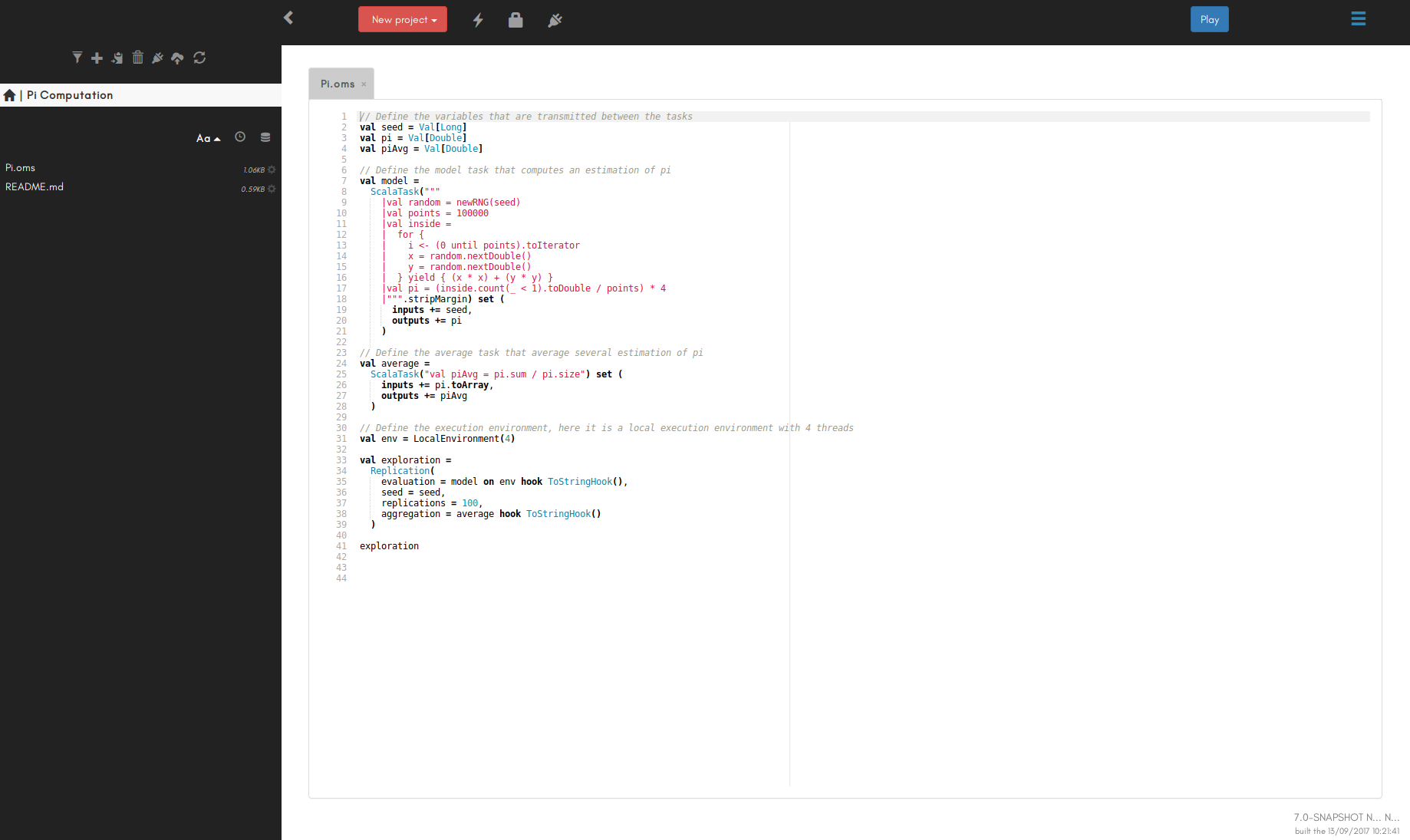
In the OpenMOLE interface, first create a file named example.oms (the file extension matters). Open it and write the following:
You can run it by clicking on the execution button (see the GUI guide)
.
// Define the variables that are transmitted between the tasks
val i = Val[Double]
val res = Val[Double]
// Define the model, here it is a simple task executing "res = i * 2", but it can be your model
val model =
ScalaTask("val res = i * 2") set (
inputs += i,
outputs += (i, res)
)
// Define the execution environment, here it is a local execution environment with 4 threads but
// it could be a remote cluster or a grid. The exploration will work the same way whatever the environment
// and no installation step is required on the execution environment.
val env = LocalEnvironment(4)
// Define the design of experiment
DirectSampling(
evaluation = model on env hook ToStringHook(),
sampling = i in (0.0 to 100.0 by 1.0)
)To distribute the exact same execution to a remote cluster, just switch the environment definition. The only configuration step required for this to work is to have an account on the cluster and to provide your authentication to OpenMOLE. Configuring authentications is explained in the section dedicated to each environment.
val env = SLURMEnvironment("myLogin", "cluster.iscpif.fr")
DirectSampling(
evaluation = model on env hook ToStringHook() on env,
sampling = i in (0.0 to 100.0 by 1.0)
)
This was just a tiny bit of what OpenMOLE can achieve! Now you might want to discover how to:
- include your own code/program: look at the Model section,
- define your environment: look at the environment section
- describe your design of experiments: look at the workflow and sampling documentation.
To get help you are more than welcome to contact the OpenMOLE user mailing list. To do so, you should subscribe here, and then you can post your question to the list users. We're also present on the #openmole channel of the ISC-PIF's RocketChat.